Downloads
Download


This work is licensed under a Creative Commons Attribution 4.0 International License.
Review
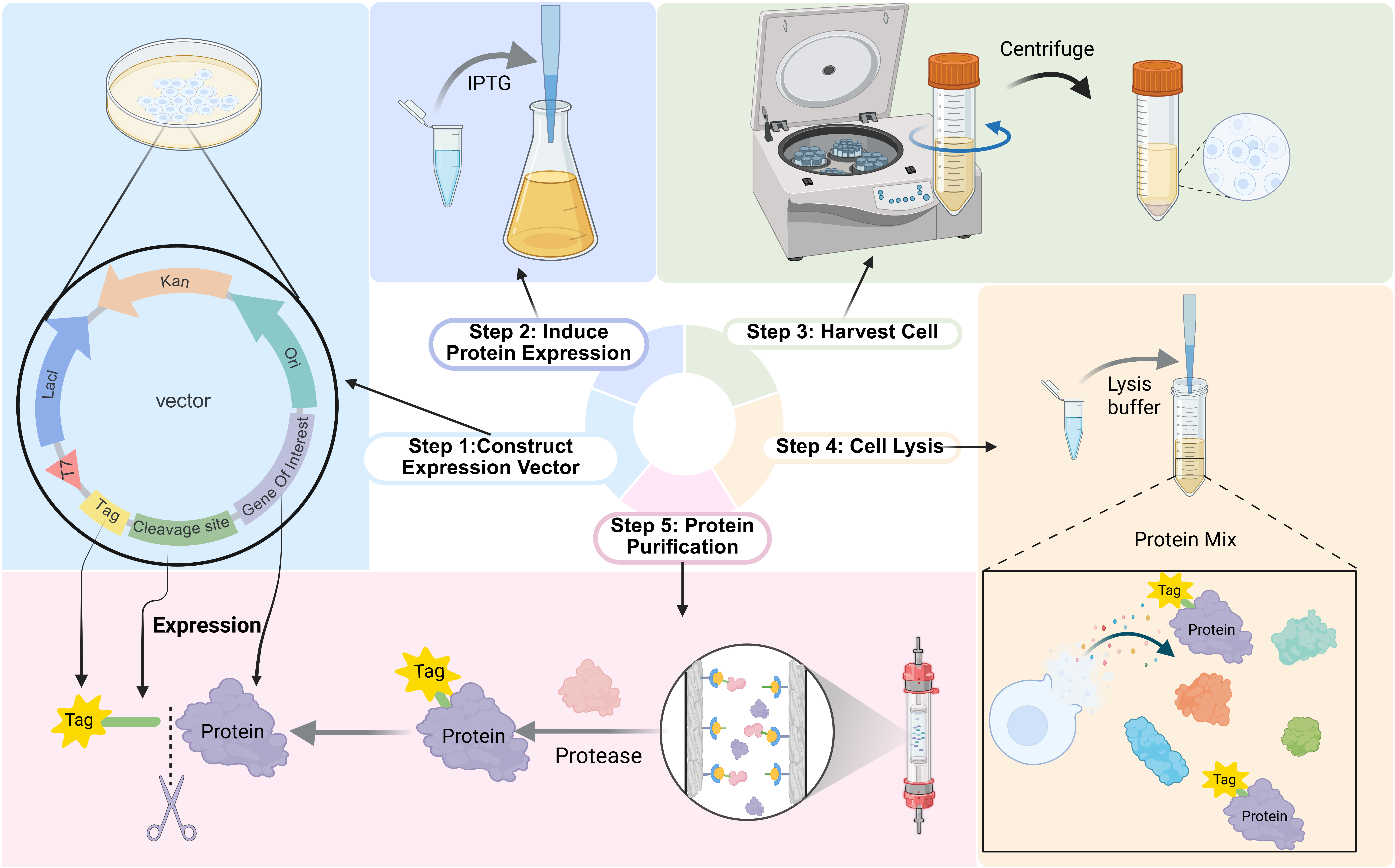
Strategies for Tag Design and Removal in the Expression and Purification of Recombinant Proteins
Jiayi Xie 1,2, Hongyi Fan 3, and Qingshan Bill Fu 1,2,*
1 School of Pharmaceutical Sciences, Southern Medical University, Guangzhou 510515, China
2 Shanghai Institute of Materia Medica, Zhongshan Institute for Drug Discovery, Chinese Academy of Sciences, Zhongshan 528400, China
3 School of Pharmacy, Guangdong Pharmaceutical University, Guangzhou 511400, China
* Correspondence: fuqingshan@simm.ac.cn
Received: 19 December 2024; Revised: 10 January 2025; Accepted: 14 February 2025; Published: 8 April 2025
Abstract: Recombinant proteins find extensive applications in the biomedical and industrial fields, and efficient protein purification is often critical for achieving their functional value. Adding specific tags to the target proteins significantly enhances expression and purification efficiency and reduces time and costs. Tags can be classified into interfering and non-interfering tags, based on their effect on protein function during purification. However, interfering tags may need to be removed after purification to prevent interference with the protein’s function in downstream applications, presenting challenges for the design and utilization of tagged fusion proteins. In this article, we discuss the recent advancements in solubility tags and controllable aggregation tags, which have emerged as powerful tools to improve purification efficiency and address these challenges. We further outline strategies for optimal tag design and on-demand cleavage, and emphasize emerging trends, technical features, and forthcoming challenges that are shaping the future of tagged fusion protein production.
Keywords:
recombinant proteins protein purification solubility tags aggregating tags tag removal proteases self-cleavage tagsReferences
- Marzano, V.; Tilocca, B.; Fiocchi, A.G.; Vernocchi, P.; Mortera, S.L.; Urbani, A.; Roncada, P.; Putignani, L. Perusal of Food Allergens Analysis by Mass Spectrometry-Based Proteomics. J. Proteom. 2020, 215, 103636. https://doi.org/10.1016/j.jprot.2020.103636.
- Rostami-Nejad, M.; Rezaei-Tavirani, M.; Mansouri, V.; Akbari, Z.; Abdi, S. Impact of Proteomics Investigations on Gastric Cancer Treatment and Diagnosis. Gastroenterol. Hepatol. Bed Bench 2019, 12 (Suppl. 1), S1–S7.
- Wert, K.J.; Velez, G.; Kanchustambham, V.L.; Shankar, V.; Evans, L.P.; Sengillo, J.D.; Zare, R.N.; Bassuk, A.G.; Tsang, S.H.; Mahajan, V.B. Metabolite Therapy Guided by Liquid Biopsy Proteomics Delays Retinal Neurodegeneration. EBioMedicine 2020, 52, 102636. https://doi.org/10.1016/j.ebiom.2020.102636.
- Mahmoudi Gomari, M.; Saraygord-Afshari, N.; Farsimadan, M.; Rostami, N.; Aghamiri, S.; Farajollahi, M.M. Opportunities and Challenges of the Tag-Assisted Protein Purification Techniques: Applications in the Pharmaceutical Industry. Biotechnol. Adv. 2020, 45, 107653. https://doi.org/10.1016/j.biotechadv.2020.107653.
- Loughran, S.T.; Walls, D. Tagging Recombinant Proteins to Enhance Solubility and Aid Purification. Methods Mol. Biol. 2017, 1485, 131–156. https://doi.org/10.1007/978-1-4939-6412-3_8.
- Terpe, K. Overview of Bacterial Expression Systems for Heterologous Protein Production: From Molecular and Biochemical Fundamentals to Commercial Systems. Appl. Microbiol. Biotechnol. 2006, 72, 211–222. https://doi.org/10.1007/s00253-006-0465-8.
- Fu, Q.; Piai, A.; Chen, W.; Xia, K.; Chou, J.J. Structure Determination Protocol for Transmembrane Domain Oligomers. Nat. Protoc. 2019, 14, 2483–2520. https://doi.org/10.1038/s41596-019-0188-9.
- Eche, S.; Gordon, M.L. Recombinant Expression of HIV-1 Protease Using Soluble Fusion Tags in Escherichia Coli: A Vital Tool for Functional Characterization of HIV-1 Protease. Virus Res. 2021, 295, 198289. https://doi.org/10.1016/j.virusres.2020.198289.
- Esposito, D.; Chatterjee, D.K. Enhancement of Soluble Protein Expression through the Use of Fusion Tags. Curr. Opin. Biotechnol. 2006, 17, 353–358. https://doi.org/10.1016/j.copbio.2006.06.003.
- Hu, L.; Guan, C.; Zhao, Y.; Zhang, W.; Chai, R.; Teng, J.; Tian, Q.; Xun, M.; Wu, F. Cloning, Sequencing, Expression, and Purification of Aspartic Proteases Isolated from Two Human Demodex Species. Int. J. Biol. Macromol. 2023, 253 Pt 7, 127404. https://doi.org/10.1016/j.ijbiomac.2023.127404.
- de Groot, N.S.; Ventura, S. Protein Activity in Bacterial Inclusion Bodies Correlates with Predicted Aggregation Rates. J. Biotechnol. 2006, 125, 110–113. https://doi.org/10.1016/j.jbiotec.2006.02.026.
- Rinas, U.; Garcia-Fruitós, E.; Corchero, J.L.; Vázquez, E.; Seras-Franzoso, J.; Villaverde, A. Bacterial Inclusion Bodies: Discovering Their Better Half. Trends Biochem. Sci. 2017, 42, 726–737. https://doi.org/10.1016/j.tibs.2017.01.005.
- Krauss, U.; Jäger, V.D.; Diener, M.; Pohl, M.; Jaeger, K.-E. Catalytically-Active Inclusion Bodies—Carrier-Free Protein Immobilizates for Application in Biotechnology and Biomedicine. J. Biotechnol. 2017, 258, 136–147. https://doi.org/10.1016/j.jbiotec.2017.04.033.
- Ma, J.; Liu, P.; Cai, S.; Wu, T.; Chen, D.; Zhu, C.; Li, S. Discovery and Identification of a Novel Tag of HlyA60 for Protein Active Aggregate Formation in Escherichia Coli. J. Agric. Food Chem. 2024, 72, 493–503. https://doi.org/10.1021/acs.jafc.3c05860.
- Li, X.; Huang, J.; Zhou, J.; Sun, C.; Zheng, Y.; Wang, Y.; Zhu, J.; Wang, S. Acyl Carrier Protein Tag Can Enhance Tobacco Etch Virus Protease Stability and Promote Its Covalent Immobilisation. Appl. Microbiol. Biotechnol. 2023, 107, 1697–1705. https://doi.org/10.1007/s00253-023-12377-8.
- Booth, W.T.; Schlachter, C.R.; Pote, S.; Ussin, N.; Mank, N.J.; Klapper, V.; Offermann, L.R.; Tang, C.; Hurlburt, B.K.; Chruszcz, M. Impact of an N-Terminal Polyhistidine Tag on Protein Thermal Stability. ACS Omega 2018, 3, 760–768. https://doi.org/10.1021/acsomega.7b01598.
- Terpe, K. Overview of Tag Protein Fusions: From Molecular and Biochemical Fundamentals to Commercial Systems. Appl. Microbiol. Biotechnol. 2003, 60, 523–533. https://doi.org/10.1007/s00253-002-1158-6.
- Smith, D.B.; Johnson, K.S. Single-Step Purification of Polypeptides Expressed in Escherichia Coli as Fusions with Glutathione S-Transferase. Gene 1988, 67, 31–40. https://doi.org/10.1016/0378-1119(88)90005-4.
- Zhou, L.; Liu, Z.; Xu, G.; Li, L.; Xuan, K.; Xu, Y.; Zhang, R. Expression of Melittin in Fusion with GST in Escherichia Coli and Its Purification as a Pure Peptide with Good Bacteriostatic Efficacy. ACS Omega 2020, 5, 9251–9258. https://doi.org/10.1021/acsomega.0c00085.
- Antoniou, G.; Papakyriacou, I.; Papaneophytou, C. Optimization of Soluble Expression and Purification of Recombinant Human Rhinovirus Type-14 3C Protease Using Statistically Designed Experiments: Isolation and Characterization of the Enzyme. Mol. Biotechnol. 2017, 59, 407–424. https://doi.org/10.1007/s12033-017-0032-9.
- Wu, Y.; Ma, Y.; Li, L.; Yang, X. Molecular Modification, Expression and Purification of New Subtype Antioxidant Peptide from Pinctada Fucata by Recombinant Escherichia Coli to Improve Antioxidant-Activity. J. Food Sci. Technol. 2018, 55, 4266–4275. https://doi.org/10.1007/s13197-018-3365-x.
- Williams, R.M.; Harvey, J.D.; Budhathoki-Uprety, J.; Heller, D.A. Glutathione-S-Transferase Fusion Protein Nanosensor. Nano Lett. 2020, 20, 7287–7295. https://doi.org/10.1021/acs.nanolett.0c02691.
- Qin, Y.; Zheng, Z.; Chu, B.; Kong, Q.; Ke, M.; Voss, C.; Li, S.S.C.; Tian, R. Generic Plug-and-Play Strategy for High-Throughput Analysis of PTM-Mediated Protein Complexes. Anal. Chem. 2022, 94, 6799–6808. https://doi.org/10.1021/acs.analchem.2c00521.
- Du, Y.; Kobashigawa, Y.; Okazaki, K.; Ogawa, M.; Kawaguchi, T.; Sato, T.; Morioka, H. Structure-Based Design, Biophysical Characterization, and Biochemical Application of the Heterodimeric Affinity Purification Tag Based on the Schistosoma Japonicum Glutathione-S-Transferase (SjGST) Homodimer. J. Biochem. 2024, 176, 69–80. https://doi.org/10.1093/jb/mvae028.
- Nagy, S.K.; Kállai, B.M.; András, J.; Mészáros, T. A Novel Family of Expression Vectors with Multiple Affinity Tags for Wheat Germ Cell-Free Protein Expression. BMC Biotechnol. 2020, 20, 17. https://doi.org/10.1186/s12896-020-00610-5.
- Kim, H.-J.; Lee, J.-H.; Lee, K.B.; Shin, J.-W.; Kwon, M.; Lee, S.; Jeong, E.M.; Cho, S.-Y.; Kim, I.-G. Transglutaminase 2 Crosslinks the Glutathione S-Transferase Tag, Impeding Protein–Protein Interactions of the Fused Protein. Exp. Mol. Med. 2021, 53, 115–124. https://doi.org/10.1038/s12276-020-00549-9.
- Al Kashgry, N.A.T.; Abulreesh, H.H.; El-Sheikh, I.A.; Almaroai, Y.A.; Salem, R.; Mohamed, I.; Waly, F.R.; Osman, G.; Mohamed, M.S.M. Utilization of a Recombinant Defensin from Maize (Zea mays L.) as a Potential Antimicrobial Peptide. AMB Express 2020, 10, 208. https://doi.org/10.1186/s13568-020-01146-9.
- Spurlino, J.C.; Lu, G.Y.; Quiocho, F.A. The 2.3-A Resolution Structure of the Maltose- or Maltodextrin-Binding Protein, a Primary Receptor of Bacterial Active Transport and Chemotaxis. J. Biol. Chem. 1991, 266, 5202–5219. https://doi.org/10.2210/pdb1mbp/pdb.
- Riggs, P. Expression and Purification of Recombinant Proteins by Fusion to Maltose-Binding Protein. Mol. Biotechnol. 2000, 15, 51–63. https://doi.org/10.1385/MB:15:1:51.
- Kapust, R.B.; Waugh, D.S. Escherichia Coli Maltose-Binding Protein Is Uncommonly Effective at Promoting the Solubility of Polypeptides to Which It Is Fused. Protein Sci. Publ. Protein Soc. 1999, 8, 1668–1674. https://doi.org/10.1110/ps.8.8.1668.
- Vagenende, V.; Yap, M.G.S.; Trout, B.L. Mechanisms of Protein Stabilization and Prevention of Protein Aggregation by Glycerol. Biochemistry 2009, 48, 11084–11096. https://doi.org/10.1021/bi900649t.
- Routzahn, K.M.; Waugh, D.S. Differential Effects of Supplementary Affinity Tags on the Solubility of MBP Fusion Proteins. J. Struct. Funct. Genom. 2002, 2, 83–92. https://doi.org/10.1023/A:1020424023207.
- Needle, D.; Waugh, D.S. Rescuing Aggregation-Prone Proteins in Escherichia Coli with a Dual His₆-MBP Tag. Methods Mol. Biol. 2014, 1177, 81–94. https://doi.org/10.1007/978-1-4939-1034-2_7.
- Han, Y.; Guo, W.; Su, B.; Guo, Y.; Wang, J.; Chu, B.; Yang, G. High-Level Expression of Soluble Recombinant Proteins in Escherichia Coli Using an HE-Maltotriose-Binding Protein Fusion Tag. Protein Expr. Purif. 2018, 142, 25–31. https://doi.org/10.1016/j.pep.2017.09.013.
- Lénon, M.; Ke, N.; Ren, G.; Meuser, M.E.; Loll, P.J.; Riggs, P.; Berkmen, M. A Useful Epitope Tag Derived from Maltose Binding Protein. Protein Sci. Publ. Protein Soc. 2021, 30, 1235–1246. https://doi.org/10.1002/pro.4088.
- Bokhove, M.; Sadat Al Hosseini, H.; Saito, T.; Dioguardi, E.; Gegenschatz-Schmid, K.; Nishimura, K.; Raj, I.; De Sanctis, D.; Han, L.; Jovine, L. Easy Mammalian Expression and Crystallography of Maltose-Binding Protein-Fused Human Proteins. J. Struct. Biol. 2016, 194, 1–7. https://doi.org/10.1016/j.jsb.2016.01.016.
- Kim, D.S.; Kim, S.W.; Song, J.M.; Kim, S.Y.; Kwon, K.-C. A New Prokaryotic Expression Vector for the Expression of Antimicrobial Peptide Abaecin Using SUMO Fusion Tag. BMC Biotechnol. 2019, 19, 13. https://doi.org/10.1186/s12896-019-0506-x.
- Guo, L.; Chen, X.; Zeng, H.; Tian, N.; Lu, W.; Zhang, J.; Xiao, Y. Production of Recombinant Human Long-Acting IL-18 Binding Protein: Inhibitory Effect on Ulcerative Colitis in Mice. Appl. Microbiol. Biotechnol. 2023, 107, 7135–7150. https://doi.org/10.1007/s00253-023-12806-8.
- Lau, Y.-T.K.; Baytshtok, V.; Howard, T.A.; Fiala, B.M.; Johnson, J.M.; Carter, L.P.; Baker, D.; Lima, C.D.; Bahl, C.D. Discovery and Engineering of Enhanced SUMO Protease Enzymes. J. Biol. Chem. 2018, 293, 13224–13233. https://doi.org/10.1074/jbc.RA118.004146.
- Heinrich, J.; Drewniok, C.; Neugebauer, E.; Kellner, H.; Wiegert, T. The YoaW Signal Peptide Directs Efficient Secretion of Different Heterologous Proteins Fused to a StrepII-SUMO Tag in Bacillus Subtilis. Microb. Cell Factories 2019, 18, 31. https://doi.org/10.1186/s12934-019-1078-0.
- Park, A.R.; Kim, S.W.; Kim, S.Y.; Kwon, K.-C. Expression of Antimicrobial Peptide (AMP), Cecropin B, in a Fused Form to SUMO Tag with or Without Three-Glycine Linker in Escherichia Coli and Evaluation of Bacteriolytic Activity of the Purified AMP. Probiotics Antimicrob. Proteins 2021, 13, 1780–1789. https://doi.org/10.1007/s12602-021-09797-1.
- Peroutka, R.J.; Elshourbagy, N.; Piech, T.; Butt, T.R. Enhanced Protein Expression in Mammalian Cells Using Engineered SUMO Fusions: Secreted Phospholipase A2. Protein Sci. 2008, 17, 1586–1595. https://doi.org/10.1110/ps.035576.108.
- Liu, L.; Spurrier, J.; Butt, T.R.; Strickler, J.E. Enhanced Protein Expression in the Baculovirus/Insect Cell System Using Engineered SUMO Fusions. Protein Expr. Purif. 2008, 62, 21–28. https://doi.org/10.1016/j.pep.2008.07.010.
- Mohammadi, M.; Taheri, R.A.; Bemani, P.; Hashemzadeh, M.S.; Farnoosh, G.; Amini, R. Utilization of SUMO Tag and Freeze-Thawing Method for a High-Level Expression and Solubilization of Recombinant Human Angiotensinconverting Enzyme 2 (rhACE2) Protein in E. Coli. Protein Pept. Lett. 2022, 29, 605–610. https://doi.org/10.2174/0929866529666220715101357.
- Khalilvand, A.B.; Aminzadeh, S.; Sanati, M.H.; Mahboudi, F. Media Optimization for SHuffle T7 Escherichia Coli Expressing SUMO-Lispro Proinsulin by Response Surface Methodology. BMC Biotechnol. 2022, 22, 1. https://doi.org/10.1186/s12896-021-00732-4.
- Aer, L.; Jiang, Q.; Gul, I.; Qi, Z.; Feng, J.; Tang, L. Overexpression and Kinetic Analysis of Ideonella Sakaiensis PETase for Polyethylene Terephthalate (PET) Degradation. Environ. Res. 2022, 212, 113472. https://doi.org/10.1016/j.envres.2022.113472.
- Hara, K.Y.; Yagi, S.; Hirono-Hara, Y.; Kikukawa, H. A Method of Solubilizing and Concentrating Astaxanthin and Other Carotenoids. Mar. Drugs 2021, 19, 462. https://doi.org/10.3390/md19080462.
- Davis, G.D.; Elisee, C.; Newham, D.M.; Harrison, R.G. New Fusion Protein Systems Designed to Give Soluble Expression in Escherichia Coli. Biotechnol. Bioeng. 1999, 65, 382–388. https://doi.org/10.1002/(SICI)1097-0290(19991120)65:4<382::AID-BIT2>3.0.CO;2-I.
- Yang, F.; Pan, Y.; Chen, Y.; Tan, S.; Jin, M.; Wu, Z.; Huang, J. Expression and Purification of Canis Interferon α in Escherichia Coli Using Different Tags. Protein Expr. Purif. 2015, 115, 76–82. https://doi.org/10.1016/j.pep.2015.07.007.
- Smith, M.A.; Gonzalez, J.; Hussain, A.; Oldfield, R.N.; Johnston, K.A.; Lopez, K.M. Overexpression of Soluble Recombinant Human Lysyl Oxidase by Using Solubility Tags: Effects on Activity and Solubility. Enzym. Res. 2016, 2016, 5098985. https://doi.org/10.1155/2016/5098985.
- Devi, N.; Adivitiya; Khasa, Y.P. A Combinatorial Approach of N-Terminus Blocking and Codon Optimization Strategies to Enhance the Soluble Expression of Recombinant hIL-7 in E. Coli Fed-Batch Culture. Appl. Microbiol. Biotechnol. 2016, 100, 9979–9994. https://doi.org/10.1007/s00253-016-7683-5.
- Li, S.; Wang, Y.; Jiang, H.; Bai, Y.; Chen, T.; Chen, M.; Ma, M.; Yang, S.; Wu, Y.; Shi, C.; et al. Display of CCL21 on Cancer Cell Membrane through Genetic Modification Using a pH Low Insertion Peptide. Int. J. Biol. Macromol. 2023, 240, 124324. https://doi.org/10.1016/j.ijbiomac.2023.124324.
- Rasooli, F.; Hashemi, A. Efficient Expression of EpEX in the Cytoplasm of Escherichia Coli Using Thioredoxin Fusion Protein. Res. Pharm. Sci. 2019, 14, 554. https://doi.org/10.4103/1735-5362.272564.
- Schenkel, M.; Treff, A.; Deber, C.M.; Krainer, G.; Schlierf, M. Heat Treatment of Thioredoxin Fusions Increases the Purity of α-Helical Transmembrane Protein Constructs. Protein Sci. 2021, 30, 1974–1982. https://doi.org/10.1002/pro.4150.
- Tsuboi, S.; Jin, T. Recombinant Protein (Luciferase-IgG Binding Domain) Conjugated Quantum Dots for BRET-Coupled Near-Infrared Imaging of Epidermal Growth Factor Receptors. Bioconjugate Chem. 2018, 29, 1466–1474. https://doi.org/10.1021/acs.bioconjchem.8b00149.
- Zhou, P.; Lugovskoy, A.A.; Wagner, G. A Solubility-Enhancement Tag (SET) for NMR Studies of Poorly Behaving Proteins. J. Biomol. NMR 2001, 20, 11–14. https://doi.org/10.1023/A:1011258906244.
- Izmailov, S.A.; Rabdano, S.O.; Hasanbasri, Z.; Podkorytov, I.S.; Saxena, S.; Skrynnikov, N.R. Structural and Dynamic Origins of ESR Lineshapes in Spin-Labeled GB1 Domain: The Insights from Spin Dynamics Simulations Based on Long MD Trajectories. Sci. Rep. 2020, 10, 957. https://doi.org/10.1038/s41598-019-56750-y.
- Rollins, N.J.; Brock, K.P.; Poelwijk, F.J.; Stiffler, M.A.; Gauthier, N.P.; Sander, C.; Marks, D.S. Inferring Protein 3D Structure from Deep Mutation Scans. Nat. Genet. 2019, 51, 1170–1176. https://doi.org/10.1038/s41588-019-0432-9.
- Soranno, A.; Cabassi, F.; Orselli, M.E.; Cellmer, T.; Gori, A.; Longhi, R.; Buscaglia, M. Dynamics of Structural Elements of GB1 β-Hairpin Revealed by Tryptophan–Cysteine Contact Formation Experiments. J. Phys. Chem. B 2018, 122, 11468–11477. https://doi.org/10.1021/acs.jpcb.8b07399.
- Song, S.-J.; Diao, H.-P.; Moon, B.; Yun, A.; Hwang, I. The B1 Domain of Streptococcal Protein G Serves as a Multi-Functional Tag for Recombinant Protein Production in Plants. Front. Plant Sci. 2022, 13, 878677. https://doi.org/10.3389/fpls.2022.878677.
- Santo, A.A.E.; Feliciano, G.T. Genetic Algorithms Applied to Thermodynamic Rational Design of Mimetic Antibodies Based on the GB1 Domain of Streptococcal Protein G: An Atomistic Simulation Study. J. Phys. Chem. B 2021, 125, 7985–7996. https://doi.org/10.1021/acs.jpcb.1c03324.
- Hwang, P.M.; Pan, J.S.; Sykes, B.D. Targeted Expression, Purification, and Cleavage of Fusion Proteins from Inclusion Bodies in Escherichia coli. FEBS Lett. 2014, 588, 247–252. https://doi.org/10.1016/j.febslet.2013.09.028.
- Zhang, Y.; Xie, X.-Q. Biosynthesis, Purification, and Characterization of a Cannabinoid Receptor 2 Fragment (CB2(271-326)). Protein Expr. Purif. 2008, 59, 249–257. https://doi.org/10.1016/j.pep.2008.02.005.
- Tang, M.; Cao, R.; Du, L.; Xu, J.; Wu, B.; OuYang, B. Ni2+ Catalyzed Cleavage of TrpLE-Fused Small Transmembrane Peptides. ChemBioChem 2022, 23, e202100514. https://doi.org/10.1002/cbic.202100514.
- Xie, X.-Q.; Zhao, J.; Zheng, H. Expression, Purification, and Isotope Labeling of Cannabinoid CB2 Receptor Fragment, CB2(180-233). Protein Expr. Purif. 2004, 38, 61–68. https://doi.org/10.1016/j.pep.2004.07.020.
- Reuten, R.; Nikodemus, D.; Oliveira, M.B.; Patel, T.R.; Brachvogel, B.; Breloy, I.; Stetefeld, J.; Koch, M. Maltose-Binding Protein (MBP), a Secretion-Enhancing Tag for Mammalian Protein Expression Systems. PLoS ONE 2016, 11, e0152386. https://doi.org/10.1371/journal.pone.0152386.
- Andler, R.; Heger, F.; Andreeßen, C.; Steinbüchel, A. Enhancing the Synthesis of Latex Clearing Protein by Different Cultivation Strategies. J. Biotechnol. 2019, 297, 32–40. https://doi.org/10.1016/j.jbiotec.2019.03.019.
- Jugniot, N.; Bam, R.; Paulmurugan, R. Expression and Purification of a Native Thy1-Single-Chain Variable Fragment for Use in Molecular Imaging. Sci. Rep. 2021, 11, 23026. https://doi.org/10.1038/s41598-021-02445-2.
- Hu, S.; Chen, X.; Zhu, M.; Hu, H.; Zhou, C.; Shi, L.; Zhang, N. Expression, Purification and Characterization of the Second DUSP Domain of Deubiquitinase USP20/VDU2. Protein Expr. Purif. 2021, 181, 105836. https://doi.org/10.1016/j.pep.2021.105836.
- Agboigba, E.; Kuchaev, E.; Garaeva, N.; Klochkova, E.; Varfolomeev, A.; Usachev, K.; Yusupov, M.; Validov, S. ORF19.2286 Gene: Isolation and Purification of Deoxyhypusine Hydroxylase from the Human Pathogenic Yeast Candida albicans. Mol. Biol. 2022, 56, 323–324. https://doi.org/10.31857/S0026898422020021.
- Zou, Z.; Cao, L.; Zhou, P.; Su, Y.; Sun, Y.; Li, W. Hyper-acidic protein fusion partners improve solubility and assist correct folding of recombinant proteins expressed in Escherichia coli. J. Biotechnol. 2008, 135, 333–339. https://doi.org/10.1016/j.jbiotec.2008.05.007.
- Yang, Y.; Ke, Z.; Wang, Z.; Li, Y.; Li, Y.; Wang, Y.; Liu, B. 1H, 13C and 15N NMR Assignments of Solubility Tag Protein Msyb of Escherichia coli. Biomol. NMR Assign. 2020, 14, 251–254. https://doi.org/10.1007/s12104-020-09955-6.
- Nguyen, T.K.M.; Ki, M.R.; Son, R.G.; Pack, S.P. The NT11, a Novel Fusion Tag for Enhancing Protein Expression in Escherichia coli. Appl. Microbiol. Biotechnol. 2019, 103, 2205–2216. https://doi.org/10.1007/s00253-018-09595-w.
- Zou, L.; Li, S.; Li, N.; Ruan, S.-L.; Chen, J.; Wu, J.; Yan, D.; Chao, H.-J. The Protocatechuate 3,4-Dioxygenase Solubility (PCDS) Tag Enhances the Expression and Solubility of Heterogenous Proteins in Escherichia coli. Front. Microbiol. 2021, 12, 779541. https://doi.org/10.3389/fmicb.2021.779541.
- Hwang, I.S.; Kim, J.H.; Jo, B.H. Enhanced Production of a Thermostable Carbonic Anhydrase in Escherichia Coli by Using a Modified NEXT Tag. Molecules 2021, 26, 5830. https://doi.org/10.3390/molecules26195830.
- Jo, B.H. An Intrinsically Disordered Peptide Tag That Confers an Unusual Solubility to Aggregation-Prone Proteins. Appl. Environ. Microbiol. 2022, 88, e0009722. https://doi.org/10.1128/aem.00097-22.
- Kronqvist, N.; Sarr, M.; Lindqvist, A.; Nordling, K.; Otikovs, M.; Venturi, L.; Pioselli, B.; Purhonen, P.; Landreh, M.; Biverstål, H.; et al. Efficient Protein Production Inspired by How Spiders Make Silk. Nat. Commun. 2017, 8, 15504. https://doi.org/10.1038/ncomms15504.
- Parida, P.P.; Saraswathi, D.; Mopidevi, S.M.V.; Raran-Kurussi, S. Advancing Large-Scale Production of Tev Protease Through an Innovative Nt* Tag-Based Fusion Construct. Curr. Res. Struct. Biol. 2023, 6, 100106. https://doi.org/10.1016/j.crstbi.2023.100106.
- Nautiyal, K.; Kuroda, Y. A SEP Tag Enhances the Expression, Solubility and Yield of Recombinant TEV Protease without Altering Its Activity. New Biotechnol. 2018, 42, 77–84. https://doi.org/10.1016/j.nbt.2018.02.006.
- Kibria, M.G.; Fukutani, A.; Akazawa-Ogawa, Y.; Hagihara, Y.; Kuroda, Y. Anti-EGFR VHH Antibody under Thermal Stress Is Better Solubilized with a Lysine than with an Arginine SEP Tag. Biomolecules 2021, 11, 810. https://doi.org/10.3390/biom11060810.
- Brindha, S.; Kibria, Md. G.; Saotome, T.; Unzai, S.; Kuroda, Y. EGFR Extracellular Domain III Expressed in Escherichia Coli with SEP Tag Shows Improved Biophysical and Functional Properties and Generate Anti-Sera Inhibiting Cancer Cell Growth. Biochem. Biophys. Res. Commun. 2021, 555, 121–127. https://doi.org/10.1016/j.bbrc.2021.03.102.
- He, W.; Zhang, J.; Sachsenhauser, V.; Wang, L.; Bardwell, J.C.A.; Quan, S. Increased Surface Charge in the Protein Chaperone Spy Enhances Its Anti-Aggregation Activity. J. Biol. Chem. 2020, 295, 14488–14500. https://doi.org/10.1074/jbc.RA119.012300.
- Ruan, A.; Ren, C.; Quan, S. Conversion of the Molecular Chaperone Spy into a Novel Fusion Tag to Enhance Recombinant Protein Expression. J. Biotechnol. 2020, 307, 131–138. https://doi.org/10.1016/j.jbiotec.2019.11.006.
- Lin, P.-H.; Tsai, S.-T.; Chang, Y.-C.; Chou, Y.-J.; Yeh, Y.-C. Harnessing Split Fluorescent Proteins in Modular Protein Logic for Advanced Whole-Cell Detection. Anal. Chim. Acta 2023, 1275, 341593. https://doi.org/10.1016/j.aca.2023.341593.
- Sheng, Y.; Li, Z.; Lin, X.; Ma, Y.; Ren, Y.; Su, Z.; Ma, G.; Zhang, S. The Position of Spy Tag/Catcher System in Hepatitis B Core Protein Particles Affects the Immunogenicity and Stability of the Synthetic Vaccine. Vaccine 2023, 41, 4867–4878. https://doi.org/10.1016/j.vaccine.2023.06.070.
- Xie, X.; Wu, P.; Huang, X.; Bai, W.; Li, B.; Shi, N. Retro-Protein XXA Is a Remarkable Solubilizing Fusion Tag for Inclusion Bodies. Microb. Cell Factories 2022, 21, 51. https://doi.org/10.1186/s12934-022-01776-7.
- Zou, S.-P.; Han, X.; Zhu, H.-Y.; Sheng, Q.; Tang, H.; Liu, Z.-Q.; Zheng, Y.-G. Functional Expression of an Echinocandin B Deacylase from Actinoplanes Utahensis in Escherichia coli. Int. J. Biol. Macromol. 2021, 187, 850–857. https://doi.org/10.1016/j.ijbiomac.2021.07.146.
- Liu, J.; Jin, Y.; Feng, X.; Zou, S.; Lv, G.; Zhang, Z.; Yang, Z. Solubility-Enhanced gMYL6 Fused with a Hexa-Lysine Tag Promotes the Cytotoxicity of Human NK Cells. Immunol. Lett. 2018, 198, 66–73. https://doi.org/10.1016/j.imlet.2018.04.003.
- Jo, B.H. Improved Solubility and Stability of a Thermostable Carbonic Anhydrase via Fusion with Marine-Derived Intrinsically Disordered Solubility Enhancers. Int. J. Mol. Sci. 2024, 25, 1139. https://doi.org/10.3390/ijms25021139.
- Carlos Rodríguez-Cabello, J.; Reguera, J.; Girotti, A.; Alonso, M.; Testera, A.M. Developing Functionality in Elastin-like Polymers by Increasing Their Molecular Complexity: The Power of the Genetic Engineering Approach. Prog. Polym. Sci. 2005, 30, 1119–1145. https://doi.org/10.1016/j.progpolymsci.2005.07.004.
- Heidari-Japelaghi, R.; Valizadeh, M.; Haddad, R.; Dorani-Uliaie, E.; Jalali-Javaran, M. Fusion to Elastin-like Polypeptide Increases Production of Bioactive Human IFN-γ in Tobacco. Transgenic Res. 2020, 29, 381–394. https://doi.org/10.1007/s11248-020-00205-y.
- Jenkins, I.C.; Milligan, J.J.; Chilkoti, A. Genetically Encoded Elastin-Like Polypeptides for Drug Delivery. Adv. Healthc. Mater. 2021, 10, e2100209. https://doi.org/10.1002/adhm.202100209.
- Meyer, D.E.; Chilkoti, A. Purification of Recombinant Proteins by Fusion with Thermally-Responsive Polypeptides. Nat. Biotechnol. 1999, 17, 1112–1115. https://doi.org/10.1038/15100.
- Fletcher, E.E.; Yan, D.; Kosiba, A.A.; Zhou, Y.; Shi, H. Biotechnological Applications of Elastin-like Polypeptides and the Inverse Transition Cycle in the Pharmaceutical Industry. Protein Expr. Purif. 2019, 153, 114–120. https://doi.org/10.1016/j.pep.2018.09.006.
- Haas, S.; Desombre, M.; Kirschhöfer, F.; Huber, M.C.; Schiller, S.M.; Hubbuch, J. Purification of a Hydrophobic Elastin-Like Protein Toward Scale-Suitable Production of Biomaterials. Front. Bioeng. Biotechnol. 2022, 10, 878838.
- Sweet, C.; Aayush, A.; Readnour, L.; Solomon, K.V.; Thompson, D.H. Development of a Fast Organic Extraction-Precipitation Method for Improved Purification of Elastin-Like Polypeptides That Is Independent of Sequence and Molecular Weight. Biomacromolecules 2021, 22, 1990–1998. https://doi.org/10.1021/acs.biomac.1c00096.
- Peyret, J.L.; Bayan, N.; Joliff, G.; Gulik-Krzywicki, T.; Mathieu, L.; Schechter, E.; Leblon, G. Characterization of the cspB Gene Encoding PS2, an Ordered Surface-Layer Protein in Corynebacterium Glutamicum. Mol. Microbiol. 1993, 9, 97–109. https://doi.org/10.1111/j.1365-2958.1993.tb01672.x.
- Nonaka, T.; Tsurui, N.; Mannen, T.; Kikuchi, Y.; Shiraki, K. Non-Chromatographic Purification of Teriparatide with a pH-Responsive CspB Tag. Protein Expr. Purif. 2019, 155, 66–71. https://doi.org/10.1016/j.pep.2018.11.008.
- Nasu, E.; Kawakami, N.; Ohara, N.; Hayashi, K.; Miyamoto, K. Column-Free Purification of an Artificial Protein Nanocage, TIP60. Protein Expr. Purif. 2023, 205, 106232. https://doi.org/10.1016/j.pep.2023.106232.
- Oki, S.; Nonaka, T.; Shiraki, K. Specific Solubilization of Impurities in Culture Media: Arg Solution Improves Purification of pH-Responsive Tag CspB50 with Teriparatide. Protein Expr. Purif. 2018, 146, 85–90. https://doi.org/10.1016/j.pep.2018.02.001.
- Nagano, H.; Mannen, T.; Kikuchi, Y.; Shiraki, K. Solution Design to Extend the pH Range of the pH-Responsive Precipitation of a CspB Fusion Protein. Protein Expr. Purif. 2022, 195–196, 106091. https://doi.org/10.1016/j.pep.2022.106091.
- Wagner, D.E.; Phillips, C.L.; Ali, W.M.; Nybakken, G.E.; Crawford, E.D.; Schwab, A.D.; Smith, W.F.; Fairman, R. Toward the Development of Peptide Nanofilaments and Nanoropes as Smart Materials. Proc. Natl. Acad. Sci. USA 2005, 102, 12656–12661. https://doi.org/10.1073/pnas.0505871102.
- Zeng, G.; Zheng, Y.; Xiang, Y.; Liu, R.; Yang, X.; Lin, Z. A Novel Protein Purification Scheme Based on Salt Inducible Self-Assembling Peptides. Microb. Cell Factories 2023, 22, 224. https://doi.org/10.1186/s12934-023-02229-5.
- Liu, Z.; Zhao, Z.; Zeng, K.; Xia, Y.; Xu, W.; Wang, R.; Guo, J.; Xie, H. Functional Immobilization of a Biofilm-Releasing Glycoside Hydrolase Dispersin B on Magnetic Nanoparticles. Appl. Biochem. Biotechnol. 2022, 194, 737–747. https://doi.org/10.1007/s12010-021-03673-y.
- Jiang, M.; Zhang, L.; Wang, F.; Zhang, J.; Liu, G.; Gao, B.; Wei, D. Novel Application of Magnetic Protein: Convenient One-Step Purification and Immobilization of Proteins. Sci. Rep. 2017, 7, 13329. https://doi.org/10.1038/s41598-017-13648-x.
- Nonaka, T.; Tsurui, N.; Mannen, T.; Kikuchi, Y.; Shiraki, K. A New pH-Responsive Peptide Tag for Protein Purification. Protein Expr. Purif. 2018, 146, 91–96. https://doi.org/10.1016/j.pep.2018.02.004.
- Fang, K.; Li, Y.; Yin, X.; Samad, A.; Jin, T. High-Level Prokaryotic Expression and Purification of Death Domain Superfamily with MBP Tag. Clin. Lab. 2018, 64, 467–475. https://doi.org/10.7754/Clin.Lab.2017.170922.
- Reddy Patakottu, B.K.; Vedire, V.R.; Reddy, C.R. Robust Production of Active Ulp1 (SUMO Protease) from Inclusion Bodies. Protein Expr. Purif. 2023, 211, 106328. https://doi.org/10.1016/j.pep.2023.106328.
- Meng, L.; Liu, Y.; Yin, X.; Zhou, H.; Wu, J.; Wu, M.; Yang, L. Effects of His-Tag on Catalytic Activity and Enantioselectivity of Recombinant Transaminases. Appl. Biochem. Biotechnol. 2020, 190, 880–895. https://doi.org/10.1007/s12010-019-03117-8.
- Majorek, K.A.; Kuhn, M.L.; Chruszcz, M.; Anderson, W.F.; Minor, W. Double Trouble-Buffer Selection and His-Tag Presence May Be Responsible for Nonreproducibility of Biomedical Experiments. Protein Sci. Publ. Protein Soc. 2014, 23, 1359–1368. https://doi.org/10.1002/pro.2520.
- Arnau, J.; Lauritzen, C.; Petersen, G.E.; Pedersen, J. Current Strategies for the Use of Affinity Tags and Tag Removal for the Purification of Recombinant Proteins. Protein Expr. Purif. 2006, 48, 1–13. https://doi.org/10.1016/j.pep.2005.12.002.
- Damough, S.; Alizadeh, R.; Komijani, S.; Shirin, M.; Adeli, A.; Mafakher, L.; Mahboudi, F.; Talebkhan Garoosi, Y. Computational and Experimental Evaluation of Linker Peptides and Thioredoxin Fusion Tag in CD20-Rituximab Specific Interactions. Iran. J. Pharm. Res. IJPR 2022, 21, e134267. https://doi.org/10.5812/ijpr-134267.
- Soleimanpour, S.; Hassannia, T.; Motiee, M.; Amini, A.A.; Rezaee, S. a. R. Fcγ1 Fragment of IgG1 as a Powerful Affinity Tag in Recombinant Fc-Fusion Proteins: Immunological, Biochemical and Therapeutic Properties. Crit. Rev. Biotechnol. 2017, 37, 371–392. https://doi.org/10.3109/07388551.2016.1163323.
- Guo, Y.; Yu, M.; Jing, N.; Zhang, S. Production of Soluble Bioactive Mouse Leukemia Inhibitory Factor from Escherichia Coli Using MBP Tag. Protein Expr. Purif. 2018, 150, 86–91. https://doi.org/10.1016/j.pep.2018.05.006.
- Taylor, A.L.; Haze-Filderman, A.; Blumenfeld, A.; Shay, B.; Dafni, L.; Rosenfeld, E.; Leiser, Y.; Fermon, E.; Gruenbaum-Cohen, Y.; Deutsch, D. High Yield of Biologically Active Recombinant Human Amelogenin Using the Baculovirus Expression System. Protein Expr. Purif. 2006, 45, 43–53. https://doi.org/10.1016/j.pep.2005.05.010.
- Uhlén, M.; Forsberg, G.; Moks, T.; Hartmanis, M.; Nilsson, B. Fusion Proteins in Biotechnology. Curr. Opin. Biotechnol. 1992, 3, 363–369. https://doi.org/10.1016/0958-1669(92)90164-e.
- Singh, H.; Verma, D.; Bardiaux, B. Single-Site Phosphorylation within the His-Tag Sequence Attached to a Recombinant Protein. Anal. Biochem. 2019, 570, 62–64. https://doi.org/10.1016/j.ab.2019.02.001.
- Schmidt, P.M.; Sparrow, L.G.; Attwood, R.M.; Xiao, X.; Adams, T.E.; McKimm-Breschkin, J.L. Taking down the FLAG! How Insect Cell Expression Challenges an Established Tag-System. PLoS ONE 2012, 7, e37779. https://doi.org/10.1371/journal.pone.0037779.
- Oliveira, C.; Domingues, L. Guidelines to Reach High-Quality Purified Recombinant Proteins. Appl. Microbiol. Biotechnol. 2018, 102, 81–92. https://doi.org/10.1007/s00253-017-8623-8.
- Young, C.L.; Britton, Z.T.; Robinson, A.S. Recombinant Protein Expression and Purification: A Comprehensive Review of Affinity Tags and Microbial Applications. Biotechnol. J. 2012, 7, 620–634. https://doi.org/10.1002/biot.201100155.
- Lichty, J.J.; Malecki, J.L.; Agnew, H.D.; Michelson-Horowitz, D.J.; Tan, S. Comparison of Affinity Tags for Protein Purification. Protein Expr. Purif. 2005, 41, 98–105. https://doi.org/10.1016/j.pep.2005.01.019.
- Lebendiker, M.; Danieli, T. Production of Prone-to-Aggregate Proteins. FEBS Lett. 2014, 588, 236–246. https://doi.org/10.1016/j.febslet.2013.10.044.
- Pereira, A.M.; Costa, A.D.; Dias, S.C.; Casal, M.; Machado, R. Production and Purification of Two Bioactive Antimicrobial Peptides Using a Two-Step Approach Involving an Elastin-Like Fusion Tag. Pharmaceuticals 2021, 14, 956. https://doi.org/10.3390/ph14100956.
- Hu, F.; Ke, T.; Li, X.; Mao, P.H.; Jin, X.; Hui, F.L.; Ma, X.D.; Ma, L.X. Expression and Purification of an Antimicrobial Peptide by Fusion with Elastin-like Polypeptides in Escherichia coli. Appl. Biochem. Biotechnol. 2010, 160, 2377–2387. https://doi.org/10.1007/s12010-009-8850-2.
- Zerweck, J.; Venkata, B.S.; Pittman, J.M.; Srivastava, A.K.; Moore, P.C.; Sachleben, J.R.; Thinakaran, G.; Meredith, S.C. A Versatile Method for Producing Labeled or Unlabeled Aβ55, Aβ40, and Other β-Amyloid Family Peptides. Protein Expr. Purif. 2019, 162, 72–82. https://doi.org/10.1016/j.pep.2019.04.006.
- Mazloomrezaei, M.; Hosseini, M.; Ahmadi, N.; Mahmoudi Maymand, E.; Eftekhar, E.; Asgari, A.; Ramezani, A. Introducing a New Method for Purification of Human IL-4 by Substitution of a Single Amino Acid in IL-4 Protein Sequence. Iran. J. Immunol. 2022, 19, 436–445. https://doi.org/10.22034/IJI.2022.94985.2336.
- Vit, O.; Harant, K.; Klener, P.; Man, P.; Petrak, J. A Three-Pronged “Pitchfork” Strategy Enables an Extensive Description of the Human Membrane Proteome and the Identification of Missing Proteins. J. Proteom. 2019, 204, 103411. https://doi.org/10.1016/j.jprot.2019.103411.
- Carter, P. Site-Specific Proteolysis of Fusion Proteins. In Protein Purification; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 1990; Volume 427, pp 181–193. https://doi.org/10.1021/bk-1990-0427.ch013.
- Riegerová, P.; Horváth, M.; Šebesta, F.; Sýkora, J.; Šulc, M.; Vlček, A. Single-Step Purification and Characterization of Pseudomonas Aeruginosa Azurin. Protein Expr. Purif. 2024, 224, 106566. https://doi.org/10.1016/j.pep.2024.106566.
- Raducanu, V.-S.; Tehseen, M.; Shirbini, A.; Raducanu, D.-V.; Hamdan, S.M. Two Chromatographic Schemes for Protein Purification Involving the Biotin/Avidin Interaction under Native Conditions. J. Chromatogr. A 2020, 1621, 461051. https://doi.org/10.1016/j.chroma.2020.461051.
- Gwak, W.S.; Choi, J.B.; Han, B.K.; Bae, S.M.; Woo, S.D. Enhanced Production of Recombinant Protein by Fusion Expression with Ssp DnaB Mini-Intein in the Baculovirus Expression System. Viruses 2018, 10, 523. https://doi.org/10.3390/v10100523.
- Dougherty, W.G.; Carrington, J.C.; Cary, S.M.; Parks, T.D. Biochemical and Mutational Analysis of a Plant Virus Polyprotein Cleavage Site. EMBO J. 1988, 7, 1281–1287. https://doi.org/10.1002/j.1460-2075.1988.tb02942.x.
- Waugh, D.S. Making the Most of Affinity Tags. Trends Biotechnol. 2005, 23, 316–320. https://doi.org/10.1016/j.tibtech.2005.03.012.
- Hu, J.; Chen, Y.; Ren, Y.; Xiao, W.; Hu, Y.; Yu, X.; Fan, J. Combination of the Mutations for Improving Activity of TEV Protease in Inclusion Bodies. Bioprocess Biosyst. Eng. 2021, 44, 2129–2139. https://doi.org/10.1007/s00449-021-02589-5.
- Nam, H.; Hwang, B.J.; Choi, D.; Shin, S.; Choi, M. Tobacco Etch Virus (TEV) Protease with Multiple Mutations to Improve Solubility and Reduce Self-Cleavage Exhibits Enhanced Enzymatic Activity. FEBS Open Bio 2020, 10, 619–626. https://doi.org/10.1002/2211-5463.12828.
- Denard, C.A.; Paresi, C.; Yaghi, R.; McGinnis, N.; Bennett, Z.; Yi, L.; Georgiou, G.; Iverson, B.L. YESS 2.0, a Tunable Platform for Enzyme Evolution, Yields Highly Active TEV Protease Variants. ACS Synth. Biol. 2021, 10, 63–71. https://doi.org/10.1021/acssynbio.0c00452.
- Paththamperuma, C.; Page, R.C. Fluorescence Dequenching Assay for the Activity of TEV Protease. Anal. Biochem. 2022, 659, 114954. https://doi.org/10.1016/j.ab.2022.114954.
- Lu, C.; Lubin, J.H.; Sarma, V.V.; Stentz, S.Z.; Wang, G.; Wang, S.; Khare, S.D. Prediction and Design of Protease Enzyme Specificity Using a Structure-Aware Graph Convolutional Network. Proc. Natl. Acad. Sci. USA 2023, 120, e2303590120. https://doi.org/10.1073/pnas.2303590120.
- Frey, S.; Görlich, D. A New Set of Highly Efficient, Tag-Cleaving Proteases for Purifying Recombinant Proteins. J. Chromatography. A 2014, 1337, 95–105. https://doi.org/10.1016/j.chroma.2014.02.029.
- Li, S.-J.; Hochstrasser, M. A New Protease Required for Cell-Cycle Progression in Yeast. Nature 1999, 398, 246–251. https://doi.org/10.1038/18457.
- Zhou, X.-F.; Zhang, C.-L.; Gao, X.-P.; Wang, W.-L.; He, Z.-F.; Jiang, F.-Y.; Pang, Y.-L.; Li, J.-H.; Ren, X.-J.; Zhou, H.-B.; et al. A Simple and Rapid Protein Purification Method Based on Cell-Surface Display of SUMO-Fused Recombinant Protein and Ulp1 Protease. AMB Express 2020, 10, 65. https://doi.org/10.1186/s13568-020-00999-4.
- Xu, H.; Wang, Q.; Zhang, Z.; Yi, L.; Ma, L.; Zhai, C. A Simplified Method to Remove Fusion Tags from a Xylanase of Bacillus Sp. HBP8 with HRV 3C Protease. Enzym. Microb. Technol. 2019, 123, 15–20. https://doi.org/10.1016/j.enzmictec.2019.01.004.
- Fan, X.; Li, X.; Zhou, Y.; Mei, M.; Liu, P.; Zhao, J.; Peng, W.; Jiang, Z.-B.; Yang, S.; Iverson, B.L.; et al. Quantitative Analysis of the Substrate Specificity of Human Rhinovirus 3C Protease and Exploration of Its Substrate Recognition Mechanisms. ACS Chem. Biol. 2020, 15, 63–73. https://doi.org/10.1021/acschembio.9b00539.
- Mei, M.; Fan, X.; Zhou, Y.; Zhang, F.; Zhang, G.; Yi, L. A Combinatorial Strategy for HRV 3C Protease Engineering to Achieve the N-Terminal Free Cleavage. Int. J. Biol. Macromol. 2024, 265, 131066. https://doi.org/10.1016/j.ijbiomac.2024.131066.
- Dušeková, E.; Berta, M.; Sedláková, D.; Řeha, D.; Dzurillová, V.; Shaposhnikova, A.; Fadaei, F.; Tomková, M.; Minofar, B.; Sedlák, E. Specific Anion Effect on Properties of HRV 3C Protease. Biophys. Chem. 2022, 287, 106825. https://doi.org/10.1016/j.bpc.2022.106825.
- Lingg, N.; Cserjan‐Puschmann, M.; Fischer, A.; Engele, P.; Kröß, C.; Schneider, R.; Brocard, C.; Berkemeyer, M.; Striedner, G.; Jungbauer, A. Advanced Purification Platform Using Circularly Permuted Caspase-2 for Affinity Fusion-Tag Removal to Produce Native Fibroblast Growth Factor 2. J. Chem. Technol. Biotechnol. 2021, 96, 1515–1522. https://doi.org/10.1002/jctb.6666.
- Lingg, N.; Kröß, C.; Engele, P.; Öhlknecht, C.; Köppl, C.; Fischer, A.; Lier, B.; Loibl, J.; Sprenger, B.; Liu, J.; et al. CASPON Platform Technology: Ultrafast Circularly Permuted Caspase-2 Cleaves Tagged Fusion Proteins before All 20 Natural Amino Acids at the N-Terminus. New Biotechnol. 2022, 71, 37–46. https://doi.org/10.1016/j.nbt.2022.07.002.
- Cserjan-Puschmann, M.; Lingg, N.; Engele, P.; Kröß, C.; Loibl, J.; Fischer, A.; Bacher, F.; Frank, A.-C.; Öhlknecht, C.; Brocard, C.; et al. Production of Circularly Permuted Caspase-2 for Affinity Fusion-Tag Removal: Cloning, Expression in Escherichia Coli, Purification, and Characterization. Biomolecules 2020, 10, 1592. https://doi.org/10.3390/biom10121592.
- Liu, J.; Fischer, A.; Cserjan-Puschmann, M.; Lingg, N.; Oostenbrink, C. Caspase-Based Fusion Protein Technology: Substrate Cleavability Described by Computational Modeling and Simulation. J. Chem. Inf. Model. 2024, 64, 5691–5700. https://doi.org/10.1021/acs.jcim.4c00316.
- Ishida, Y.; Inouye, M. Suppression of the Toxicity of Bac7 (1–35), a Bovine Peptide Antibiotic, and Its Production in E. Coli. AMB Express 2016, 6, 19. https://doi.org/10.1186/s13568-016-0190-3.
- Huth, J.R.; Bewley, C.A.; Clore, G.M.; Gronenborn, A.M.; Jackson, B.M.; Hinnebusch, A.G. Design of an Expression System for Detecting Folded Protein Domains and Mapping Macromolecular Interactions by NMR. Protein Sci. 1997, 6, 2359–2364. https://doi.org/10.1002/pro.5560061109.
- Zlobin, A.; Golovin, A. Between Protein Fold and Nucleophile Identity: Multiscale Modeling of the TEV Protease Enzyme–Substrate Complex. ACS Omega 2022, 7, 40279–40292. https://doi.org/10.1021/acsomega.2c05201.
- Naor, A.; Altman-Price, N.; Soucy, S.M.; Green, A.G.; Mitiagin, Y.; Turgeman-Grott, I.; Davidovich, N.; Gogarten, J.P.; Gophna, U. Impact of a Homing Intein on Recombination Frequency and Organismal Fitness. Proc. Natl. Acad. Sci. USA 2016, 113, E4654–E4661. https://doi.org/10.1073/pnas.1606416113.
- Novikova, O.; Jayachandran, P.; Kelley, D.S.; Morton, Z.; Merwin, S.; Topilina, N.I.; Belfort, M. Intein Clustering Suggests Functional Importance in Different Domains of Life. Mol. Biol. Evol. 2016, 33, 783–799. https://doi.org/10.1093/molbev/msv271.
- Wood, D.W.; Camarero, J.A. Intein Applications: From Protein Purification and Labeling to Metabolic Control Methods *. J. Biol. Chem. 2014, 289, 14512–14519. https://doi.org/10.1074/jbc.R114.552653.
- Lennon, C.W.; Stanger, M.; Banavali, N.K.; Belfort, M. Conditional Protein Splicing Switch in Hyperthermophiles through an Intein-Extein Partnership. mBio 2018, 9, e02304-17. https://doi.org/10.1128/mBio.02304-17.
- Wood, D.W.; Belfort, M.; Lennon, C.W. Inteins—Mechanism of Protein Splicing, Emerging Regulatory Roles, and Applications in Protein Engineering. Front. Microbiol. 2023, 14, 1305848. https://doi.org/10.3389/fmicb.2023.1305848.
- Chong, S.; Mersha, F.B.; Comb, D.G.; Scott, M.E.; Landry, D.; Vence, L.M.; Perler, F.B.; Benner, J.; Kucera, R.B.; Hirvonen, C.A.; et al. Single-Column Purification of Free Recombinant Proteins Using a Self-Cleavable Affinity Tag Derived from a Protein Splicing Element. Gene 1997, 192, 271–281. https://doi.org/10.1016/s0378-1119(97)00105-4.
- Lin, Z.; Amesso Ndengue, P.P.; Jing, Y.; Zhao, L.; Yang, X. Facile Expression and Purification of Active Human Growth Hormone in E. Coli by a Cleavable Self-Aggregating Tag Scheme. Protein Expr. Purif. 2021, 188, 105974. https://doi.org/10.1016/j.pep.2021.105974.
- Lee, Y.-Z.; Sue, S.-C. Salt-Sensitive Intein for Large-Scale Polypeptide Production. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 621, pp 111–130. https://doi.org/10.1016/bs.mie.2019.02.024.
- Luo, H.; Hu, L.; Ma, B.; Zhao, M.; Luo, M.; Deng, Q.; Deng, S.; Ye, H.; Lin, T.; Chen, J.; et al. Molecular Dynamics Based Improvement of the Solubilizing Self-Cleavable Tag Zbasic-ΔI-CM Application in the Preparation of Recombinant Proteins in Escherichia coli Biochem. Biophys. Res. Commun. 2019, 513, 412–418. https://doi.org/10.1016/j.bbrc.2019.03.205.
- Lin, Z.; Zhao, Q.; Wang, X.; Zhou, B.; Xing, L.; Wang, J.; Pistolozzi, M.; Zhao, L.; Wang, T. Engineered pH-Inducible Intein Mtu ΔI-CM Variants with Markedly Reduced Premature Cleavage Activity. AIChE J. 2020, 66, e16806. https://doi.org/10.1002/aic.16806.
- Banki, M.R.; Gerngross, T.U.; Wood, D.W. Novel and Economical Purification of Recombinant Proteins: Intein-Mediated Protein Purification Using In Vivo Polyhydroxybutyrate (PHB) Matrix Association. Protein Sci. 2005, 14, 1387–1395. https://doi.org/10.1110/ps.041296305.
- Liu, F.; Tsai, S.-L.; Madan, B.; Chen, W. Engineering a High-Affinity Scaffold for Non-Chromatographic Protein Purification via Intein-Mediated Cleavage. Biotechnol. Bioeng. 2012, 109, 2829–2835. https://doi.org/10.1002/bit.24545.
- Tian, L.; Sun, S.S.M. A Cost-Effective ELP-Intein Coupling System for Recombinant Protein Purification from Plant Production Platform. PLoS ONE 2011, 6, e24183. https://doi.org/10.1371/journal.pone.0024183.
- Chen, Z.; Zhao, L.; Ru, J.; Yu, S.; Yu, H.; Ren, H.; Zhang, Y.; Zhang, W.; Lin, F.; Huo, Y.-X. A Novel Protein Purification Strategy Mediated by the Combination of CipA and Ssp DnaB Intein. J. Biotechnol. 2019, 301, 97–104. https://doi.org/10.1016/j.jbiotec.2019.06.002.
- Zhao, Q.; Xu, W.; Xing, L.; Lin, Z. Recombinant Production of Medium- to Large-Sized Peptides in Escherichia Coli Using a Cleavable Self-Aggregating Tag. Microb. Cell Factories 2016, 15, 136. https://doi.org/10.1186/s12934-016-0534-3.
- Guan, D.; Ramirez, M.; Chen, Z. Split Intein Mediated Ultra-Rapid Purification of Tagless Protein (SIRP). Biotechnol. Bioeng. 2013, 110, 2471–2481. https://doi.org/10.1002/bit.24913.
- Shi, C.; Meng, Q.; Wood, D.W. A Dual ELP-Tagged Split Intein System for Non-Chromatographic Recombinant Protein Purification. Appl. Microbiol. Biotechnol. 2013, 97, 829–835. https://doi.org/10.1007/s00253-012-4601-3.
- Li, Y. Self-Cleaving Fusion Tags for Recombinant Protein Production. Biotechnol. Lett. 2011, 33, 869–881. https://doi.org/10.1007/s10529-011-0533-8.
- Kapust, R.B.; Waugh, D.S. Controlled Intracellular Processing of Fusion Proteins by TEV Protease. Protein Expr. Purif. 2000, 19, 312–318. https://doi.org/10.1006/prep.2000.1251.
- Luo, X.; Li, L.; Chai, M.; Zhang, Q.; Shang, G. Escherichia Coli BL21(DE3) Chromosome-Based Controlled Intracellular Processing System for Fusion Protein Separation. J. Microbiol. Methods 2015, 114, 35–37. https://doi.org/10.1016/j.mimet.2015.04.013.
- Feng, Y.; Xu, Q.; Yang, T.; Sun, E.; Li, J.; Shi, D.; Wu, D. A Novel Self-Cleavage System for Production of Soluble Recombinant Protein in Escherichia coli Protein Expr. Purif. 2014, 99, 64–69. https://doi.org/10.1016/j.pep.2014.04.001.
- Wang, H.; Chu, Z.; Chen, C.; Cao, A.; Tong, X.; Ouyang, C.; Yuan, Q.; Wang, M.; Wu, Z.; Wang, H.; et al. Recombinant Passenger Proteins Can Be Conveniently Purified by One-Step Affinity Chromatography. PLoS ONE 2015, 10, e0143598. https://doi.org/10.1371/journal.pone.0143598.
- Kuo, D.; Nie, M.; Courey, A.J. SUMO as a Solubility Tag and In Vivo Cleavage of SUMO Fusion Proteins with Ulp1. In Protein Affinity Tags; Giannone, R.J., Dykstra, A.B., Eds.; Methods in Molecular Biology; Springe: New York, NY, USA, 2014; Volume 1177, pp 71–80. https://doi.org/10.1007/978-1-4939-1034-2_6.
- Babbal; Adivitiya; Mohanty, S.; Khasa, Y.P. Bioprocess Optimization for the Overproduction of Catalytic Domain of Ubiquitin-like Protease 1 (Ulp1) from S. Cerevisiae in E. Coli Fed-Batch Culture. Enzym. Microb. Technol. 2019, 120, 98–109. https://doi.org/10.1016/j.enzmictec.2018.10.008.
- Walker, P.A.; Leong, L.E.-C.; Ng, P.W.P.; Tan, S.H.; Waller, S.; Murphy, D.; Porter, A.G. Efficient and Rapid Affinity Purification of Proteins Using Recombinant Fusion Proteases. Nat. Biotechnol. 1994, 12, 601–605. https://doi.org/10.1038/nbt0694-601.
- Chen, Y.; Li, Q.; Yang, J.; Xie, H. Promoting Tag Removal of a MBP-Fused Integral Membrane Protein by TEV Protease. Appl. Biochem. Biotechnol. 2017, 181, 939–947. https://doi.org/10.1007/s12010-016-2260-z.
- Babbal; Mohanty, S.; Khasa, Y.P. Designing Ubiquitin-like Protease 1 (Ulp1) Based Nano Biocatalysts: A Promising Technology for SUMO Fusion Proteins. Int. J. Biol. Macromol. 2024, 255, 128258. https://doi.org/10.1016/j.ijbiomac.2023.128258.





