Downloads
Download


This work is licensed under a Creative Commons Attribution 4.0 International License.
Review
Exploring Fungi Within the Human Gut Microbiota: Obstacles, Innovations, Therapeutic Applications and Prospects Ahead
Peng Xue 1,†, Chao Luo 2,†, Jiashu Li 3, Liang Yang 3,* and Yuanyuan Ma 1,*
1 Nantong Key Laboratory of Environmental Toxicology, Department of Occupational Medicine and Environmental Toxicology, School of Public Health, Nantong University, Nantong 226019, China
2 The People’s Hospital of Rugao, Affiliated Rugao Hospital of Xinglin College, Nantong University, Nantong 226500, China
3 School of Medicine, Southern University of Science and Technology, Shenzhen 518055, China
* Correspondence: yangl@sustech.edu.cn (L.Y.); myycsd@ntu.edu.cn (Y.M.)
Received: 15 September 2024; Revised: 23 October 2024; Accepted: 23 October 2024; Published: 9 April 2025
Abstract: Fungi in the human gut microbiota participate in the maintenance of health and regulation of physiological processes. This review examines the complex role of gut fungi, highlighting challenges such as diverse fungal populations, cultivation difficulties, and knowledge gaps in their functional roles. Recent advancements in metagenomics and metabolomics have enabled innovative investigations into fungal communities, revealing their influence on host metabolism and immune responses. Future research should address the existing knowledge gaps and explore the therapeutic applications of gut fungi. Interdisciplinary collaborations and new methodologies are proposed to enhance the current understanding of fungi-host interactions, ultimately improving health outcomes and guide the development of novel treatment strategies. This review emphasizes the need to integrate fungal research into microbiome studies to provide a comprehensive understanding of gut health.
Keywords:
gut microbiota fungi metagenomics metabolomics immune regulation1. Introduction
Human gut microbiota is a complex and dynamic ecosystem composed of trillions of microorganisms, including bacteria, archaea, viruses, and fungi [1–4]. This intricate community regulates various physiological processes such as digestion, metabolism, and immune regulation [5–7]. Thus, understanding the composition and function of gut microbiota has attracted considerable attention, as they are influence human health and disease [8,9]. The gut microbiota actively participate in host interactions, influencing biological pathways and contributing to homeostasis [10–12]. Among the diverse microbial inhabitants of the gut, fungi have emerged as essential yet often overlooked components of the microbiome [13]. Historically, research has focused predominantly on bacteria; however, recent studies have uncovered important roles that fungi, particularly yeasts like Saccharomyces and Candida, play within the gut environment [14]. These fungal species can influence gut health through their metabolic activities, modulation of the immune response, and interactions with other microbial taxa [15,16]. For instance, alterations in the fungal composition of the gut microbiota have been linked to the occurrence of various health conditions, including inflammatory bowel disease, obesity, and metabolic syndrome [17,18]. Recent research, particularly by Bingzhai et al., provided important insights into the dynamics of gut fungi, particularly within immunocompromised populations, such as individuals undergoing allogeneic hematopoietic cell transplantation [19]. Their findings reveal the emerging challenges caused by fungal infections, specifically those attributed to Candida parapsilosis, even in patients receiving antifungal prophylaxis, such as micafungin. This situation highlights the phenomenon of heteroresistance, wherein a small, low-frequency subpopulation of resistant fungal cells can evade detection by standard antimicrobial susceptibility assays. This results in treatment failures among these high-risk patients, emphasizing the need for further investigation into the mechanisms of fungal resistance and their implications for the clinical management of patients [19].
Therefore, it is imperative to further investigate the role of gut fungi in the context of immune suppression and the mechanisms underlying breakthrough infections. This review discusses the current state of research on gut fungi, emphasizing their importance, roles, and interactions. We identify key obstacles limiting investigations into gut fungi, such as challenges in sampling, analytical techniques, and data interpretation. Finally, we discuss innovative approaches and future prospects for advancing our understanding of gut fungi, with focus on the clinical implications of gut fungi. As research in this area evolves, recognizing the significance of fungi among gut microbiota is crucial for comprehensively understanding their contributions to human health.
2. Composition and Diversity of Fungi in the Human Gut
The human gut is home to a rich diversity of fungal species, predominantly comprising various yeasts. Notable fungi include Saccharomyces cerevisiae, known for its role in fermenting dietary carbohydrates [20], and Candida species, which can exist in commensal and pathogenic forms [20,21]. Other genera such as Aspergillus, Penicillium, and Cladosporium have also been identified within the gut microbiota [22], though they are less frequently studied. Recent research by Yan et al. led to the development of a comprehensive catalog of 760 gut fungal species, including 69 previously unrecorded species [3]. The catalog revealed the considerable diversity and functional potential of gut fungi, as well as their effects on health and disease, particularly in conditions like inflammatory bowel disease.
Several factors affect the composition and diversity of gut fungi. Among them, dietary habits play crucial roles, with some types of foods known to promote the growth of specific fungal species. Diets high in protein, animal fats, and carbohydrates may enhance the proliferation of specific fermentative yeasts [23]. Antibiotic use also plays a significant role, as it disrupts bacterial populations, potentially allowing opportunistic fungi such as Candida to overgrow [24,25]. Environmental exposures, physical and psychological stress, and underlying health conditions can further shape the gut mycobiome [26]. Moreover, birth delivery methods, infant feeding practices, pH levels, water quality, and the presence of prebiotics and probiotics, can affect the overall fungal composition [26]. Fungal and bacterial communities within the gut often participate in complex symbiotic relationships. Some fungal species produce metabolites that promote the growth of beneficial bacteria, while bacteria can create conditions that support fungal survival [27]. This mutualism enhances nutrient cycling, improves gut barrier integrity, and contributes to overall host health. However, fungi and bacteria also compete for resources, with certain bacteria inhibiting fungal growth by producing antimicrobial compounds or substrate competition. Conversely, fungal metabolites may exert direct inhibition effect or induce the production of antimicrobial substances like antimicrobial peptide [28] to repress bacterial growth leading to the phylum-level [29] or more specific shift of microbial composition. Besides, metabolites play a significant role in the crosstalk between gut microbiota and human health. In heart failure cases, as the weakened barrier function of the bowel wall can promote microorganisms dissemination and heighten inflammation, metabolites like trimethylamine N-oxide (TMAO), which originally induce atherosclerosis through endocrine would further aggravate cardiac load [30]. Besides TMAO, bile acids (BAs), short chain fatty acids (SCFAs) and amino acids are among the most studied metabolites. Some them have beene linked to the development of carcinogenesis. Activation of NF-κB, JAK2 and STAT3 signaling pathways by Bas triggered colorectal cancer [31], while inhibiting intestinal motility [32], epithelial stability [33], and increasing SCFAs production prevented its development. Meanwhile, the biotransfer of these metabolites by gut fungus has been shown to have dual effects on immune homeostasis [34]. Overall, the composition and diversity of fungi in the human gut are shaped by multiple factors, and their interactions with various bacterial communities are complex and multifaceted, with important implications in gut health and disease.
3. Challenges
Several challenges limit research into gut fungi, with the major ones being sample collection, cultivation, and data analysis. Given that the abundance of fungi is lower compared to that of bacteria, it is challenging to obtain samples. Moreover, traditional cultivation techniques are not ideal for culturing various fungal species as they lack specific and complex growth requirements. This limitation can induce biases in the observed fungal diversity because the viability of fungi during sample processing and storage often diminishes, compromising the accuracy of analyses. The inherent complexity and diversity of the gut microbiome further complicate research efforts. Various microorganisms in the gut create a dynamic ecosystem, making it challenging to differentiate between commensal and pathogenic fungal species and understand their specific roles. Interactions among microbial groups can significantly alter their abundance and functionality, highlighting the importance of adopting integrative approaches to fully capture the multifactorial nature of these communities. Moreover, while high-throughput sequencing technologies have revolutionized microbiome research, they also pose unique challenges for studying fungi. The intricate nature of fungal genomes and genetic variation can complicate sequencing and data interpretation. Current bioinformatics tools are often designed primarily for bacterial analysis, leaving a gap in the methodologies suitable for analyzing fungal datasets. To address these challenges, robust and specialized bioinformatics pipelines are essential for accurate fungal analysis. Moreover, there are no standardized taxonomic frameworks for classifying fungi within the gut microbiome, which leads to discrepancies in species identification across studies. Promoting collaboration among researchers to develop unified taxonomic criteria is crucial for advancing the field and ensuring reproducibility in fungal microbiome studies. By addressing these challenges, researchers can deepen their understanding of gut fungi and their implications for human health.
4. Innovation and Development
The emergence of next-generation sequencing (NGS) technologies has revolutionized microbiome research, particularly in the study of gut fungi. NGS facilitates high-throughput analysis of complex microbial communities, facilitating the identification and characterization of various fungal species in the gastrointestinal tract. This advancement has significantly expanded our understanding of fungal diversity, abundance, and their functional roles in health and disease [35]. Recent studies have confirmed the regulatory effects of fungal microbiota or mycobiota on the immune system, particularly in inflammatory bowel disease. Although various alterations in mycobiota have been associated with diseases, the specific functional implications of individual fungal strains remain to be fully elucidated. A notable contribution in this area is the translational platform developed by Li et al., which integrates high-resolution mycobiota sequencing, culturomics, genomics, CRISPR-Cas9-based strain editing, and functional assays. This platform represents a significant advancement in exploring the interactions of fungal strains at both the individual strain and patient-specific levels [36]. Fungi contain the cell wall and have a low RNA content estimated to be 1–3 picograms per cell, which necessitates the use of specialized RNA extraction methods. Techniques such as single-cell RNA sequencing (scRNA-seq), including Smart-seq/C1 [37], YscRNA-seq [38], and SCRB-seq [39], allow for the analysis of hundreds of yeast cells and the identification of 1000 to 3000 genes per cell. YscRNA-seq is particularly adept at capturing the 5′ ends of transcripts for precise quantification of gene expression, while SCRB-seq facilitates studies based on cell size. These scRNA-seq technologies might provide groundbreaking approaches to investigate fungal population heterogeneity, enabling researchers to uncover the unique functional characteristics and metabolic capabilities of specific fungal species, especially those relevant to inflammatory bowel disease, such as certain strains of C. albicans associated with immune cell damage. Moreover, integrating metabolomics with sequencing data will uncover the detailed biochemical interactions between fungi and other microorganisms, which modulate their effects on host health and metabolic networks. Moreover, employing multi-omics approaches encompassing genomics, transcriptomics, proteomics, and metabolomics will provide coomprehensive understanding of the interactions within the gut microbiome [40]. Addressing the complexities of the gut mycobiome requires interdisciplinary collaboration among microbiology, bioinformatics, systems biology, and clinical sciences. Such collaborative efforts will lead to the development of innovative methodologies for studying gut fungi, which is essential for indepth understanding of gut fungi. The diverse metabolites produced by fungi significantly influence host health, immune responses, gut barrier integrity, and metabolic health [41]. In certain intestinal diseases, synchronous changes might appear between metabolites proportion and microorganism species and density, which could serve as the indicators of diseases and therapeutic target. Continued investigation into these metabolites and their clinical potential is crucial for identifying beneficial fungal strains and understanding their roles as both symbionts and potential pathogens. By leveraging technological advancements and promoting interdisciplinary collaboration, the full landscape of functional roles of gut fungi in health and disease will be identified, paving the way for the formulation of novel therapeutic strategies.
5. Conclusions and Future Directions
Changes in gut fungal composition are connected to health concerns (schematic in Figure 1 by Figdraw2.0). Gut fungi display considerable diversity and play complex roles within the microbiome, interacting with both the host and other microbial communities. Recent advancements in sequencing technologies and multi-omics approaches have shed light on the biological functions of fungi, revealing their potential effects on health through metabolic byproducts and immune modulation. However, our understanding of the specific mechanisms by which fungi influence health and disease is still limited. Future research should aim to clarify the physiological and ecological characteristics of gut fungi, exploring their life cycles, stress responses, and interactions with host biology. Examining how diet together with its metabolites transformed by fungi, genetics, and health status affect fungal communities will enhance our comprehension of the host-microbiome relationship, potentially leading to targeted dietary and therapeutic interventions for improved gut health. Moreover, the clinical implications of fungal research are promising. Identifying specific fungal signatures in the level of molecular and population which are linked to diseases could enable early diagnosis and personalized treatment strategies. The development of tailored microbiome therapies, based on individual fungal compositions, may enhance therapeutic outcomes through probiotics and dietary modifications. As the field advances, interdisciplinary collaboration and innovative research methodologies will be crucial for unlocking the therapeutic potential of gut fungi. Prioritizing in-depth studies on the biological characteristics and interactions of fungi within the gut microbiome will not only deepen scientific knowledge but also lead to novel clinical applications that improve health outcomes.
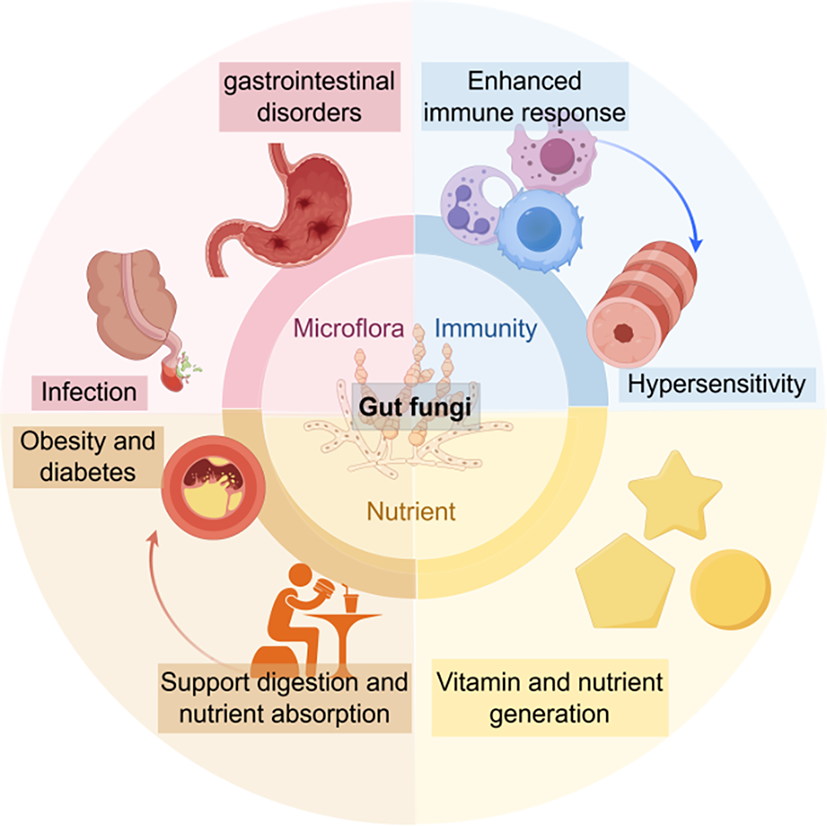
Figure 1. Schematic summary model. Gut fungi affect various aspects of gut microbiota, including microflora, immunity, nutrient production and absorption. If some originally beneficial effects exceed a certain threshold of onset, diseases may occur.
Author Contributions: P.X., C.L., and J.L.: writing—original draft preparation; L.Y. and Y.M.: writing—reviewing and editing. All authors have read and agreed to the published version of the manuscript.
Funding: This research was funded by the Natural Science Foundation of Jiangsu Province (BK20240948), Nantong Jiangsu Scientific Research Project (JC2023043), and the Natural Science Research of Jiangsu Higher Education Institutions (24KJD430010).
Institutional Review Board Statement: Not applicable.
Informed Consent Statement: Not applicable.
Data Availability Statement: Not applicable.
Conflicts of Interest: The authors declare no conflict of interest.
References
- Almeida, A.; Mitchell, A.L.; Boland, M.; et al. A new genomic blueprint of the human gut microbiota. Nature 2019, 568, 499–504.
- Leviatan, S.; Shoer, S.; Rothschild, D.; et al. An expanded reference map of the human gut microbiome reveals hundreds of previously unknown species. Nat. Commun. 2022, 13, 3863.
- Yan, Q.; Li, S.; Yan, Q.; et al. A genomic compendium of cultivated human gut fungi characterizes the gut mycobiome and its relevance to common diseases. Cell 2024, 187, 2969–2989.
- Garcia-Bonete, M.J.; Rajan, A.; Suriano, F.; et al. The underrated gut microbiota helminths, bacteriophages, fungi, and archaea. Life 2023, 13, 1765.
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71.
- Woźniak, D.; Cichy, W.; Przysławski, J.; et al. The role of microbiota and enteroendocrine cells in maintaining homeostasis in the human digestive tract. Adv. Med. Sci. 2021, 66, 284–292.
- Chu, J.; Feng, S.; Guo, C.; et al. Immunological mechanisms of inflammatory diseases caused by gut microbiota dysbiosis: A review. Biomed. Pharmacother. 2023, 164, 114985.
- Colella, M.; Charitos, I.A.; Ballini, A.; et al. Microbiota revolution: How gut microbes regulate our lives. World J. Gastroenterol. 2023, 29, 4368.
- Zhou Y.-D.; Liang F.-X.; Tian H.-R.; et al. Mechanisms of gut microbiota-immune-host interaction on glucose regulation in type 2 diabetes. Front. Microbiol. 2023, 14, 1121695.
- Fujisaka, S.; Watanabe, Y.; Tobe, K. The gut microbiome: A core regulator of metabolism. J. Endocrinol. 2023, 256, e220111.
- Kim, S.; Seo S.-U.; Kweon M.-N. Gut microbiota-derived metabolites tune host homeostasis fate. In Seminars in Immunopathology; Springer: Berlin/Heidelberg, Germany, 2024; p. 2.
- Chen, L.; Zhang, L.; Hua, H.; et al. Interactions between toll-like receptors signaling pathway and gut microbiota in host homeostasis. Immun. Inflamm. Dis. 2024, 12, e1356.
- Huseyin, C.E.; O’Toole, P.W.; Cotter, P.D.; et al. Forgotten fungi—The gut mycobiome in human health and disease. FEMS Microbiol. Rev. 2017, 41, 479–511.
- Di Paola, M.; Rizzetto, L.; Stefanini, I.; et al. Comparative immunophenotyping of Saccharomyces cerevisiae and Candida spp. strains from Crohn’s disease patients and their interactions with the gut microbiome. J. Transl. Autoimmun. 2020, 3, 100036.
- Peroumal, D.; Sahu, S.R.; Kumari, P.; et al. Commensal fungus Candida albicans maintains a long-term mutualistic relationship with the host to modulate gut microbiota and metabolism. Microbiol. Spectr. 2022, 10, e02462-22.
- Wang, X.; Wu, S.; Li, L.; et al. Candida albicans overgrowth disrupts the gut microbiota in mice bearing oral cancer. Mycology 2024, 15, 57–69.
- Pérez, J.C. Fungi of the human gut microbiota: Roles and significance. Int. J. Med. Microbiol. 2021, 311, 151490.
- Richard, M.L.; Sokol, H. The gut mycobiota: Insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 331–345.
- Zhai, B.; Liao, C.; Jaggavarapu, S.; et al. Antifungal heteroresistance causes prophylaxis failure and facilitates breakthrough Candida parapsilosis infections. Nat. Med. 2024, 30, 3163–3172.
- Bayoumy, A.B.; Mulder, C.J.J.; Mol, J.J.; et al. Gut fermentation syndrome: A systematic review of case reports. United Eur. Gastroenterol. J. 2021, 9, 332–342.
- Demir, M.; Lang, S.; Hartmann, P.; et al. The fecal mycobiome in non-alcoholic fatty liver disease. J. Hepatol. 2022, 76, 788–799.
- Hallen-Adams, H.E.; Suhr M.J. Fungi in the healthy human gastrointestinal tract. Virulence 2017, 8, 352–358.
- Liu, T.; Asif, I.M.; Chen, Y.; et al. The Relationship between Diet, Gut Mycobiome, and Functional Gastrointestinal Disorders: Evidence, Doubts, and Prospects. Mol. Nutr. Food Res. 2024, 68, e2300382.
- Kumamoto, C.A.; Gresnigt, M.S.; Hube, B. The gut, the bad and the harmless: Candida albicans as a commensal and opportunistic pathogen in the intestine. Curr. Opin. Microbiol. 2020, 56, 7–15.
- d’Enfert, C.; Kaune, A.K.; Alaban, L.R.; et al. The impact of the Fungus-Host-Microbiota interplay upon Candida albicans infections: Current knowledge and new perspectives. FEMS Microbiol. Rev. 2021, 45, fuaa060.
- Boutin, R.C.T.; Sbihi, H.; McLaughlin, R.J.; et al. Composition and Associations of the Infant Gut Fungal Microbiota with Environmental Factors and Childhood Allergic Outcomes. mBio 2021, 12, e0339620.
- Maas, E.; Penders, J. Fungal-Bacterial Interactions in the Human Gut of Healthy Individuals. J. Fungi 2023, 9, 139.
- Termén, S.; Tollin, M.; Rodriguez, E.; et al. PU.1 and bacterial metabolites regulate the human gene CAMP encoding antimicrobial peptide LL-37 in colon epithelial cells. Mol. Immunol. 2008, 45, 3947–3955.
- Islam, K.B.; Fukiya, S.; Hagio, M.; et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011, 141, 1773–1781.
- Tang, W.H.W.; Li, D.Y.; Hazen S.L. Dietary metabolism, the gut microbiome, and heart failure. Nat. Rev. Cardiol. 2019, 16, 137–154.
- Qu, R.; Zhang, Y.; Ma, Y.; et al. Role of the Gut Microbiota and Its Metabolites in Tumorigenesis or Development of Colorectal Cancer. Adv. Sci. 2023, 10, e2205563.
- Chen, J.; Vitetta, L. Intestinal dysbiosis in celiac disease: Decreased butyrate production may facilitate the onset of the disease. Proc. Natl. Acad. Sci. USA 2021, 118, e2113655118.
- Waclawiková B.; Codutti, A.; Alim, K.; et al. Gut microbiota-motility interregulation: Insights from in vivo, ex vivo and in silico studies. Gut Microbes 2022, 14, 1997296.
- Wang, J.; Zhu, N.; Su, X.; et al. Gut-Microbiota-Derived Metabolites Maintain Gut and Systemic Immune Homeostasis. Cells 2023, 12, 793.
- Gao, B.; Chi, L.; Zhu, Y. An Introduction to Next Generation Sequencing Bioinformatic Analysis in Gut Microbiome Studies. Biomolecules 2021, 11, 530.
- Li, X.V.; Leonardi, I. Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature 2022, 603, 672–678. Erratum in Nature 2022, 608, E21.
- Gasch, A.P.; Yu, F.B.; Hose, J.; et al. Single-cell RNA sequencing reveals intrinsic and extrinsic regulatory heterogeneity in yeast responding to stress. PLoS Biol. 2017, 15, e2004050.
- Nadal-Ribelles, M.; Islam, S.; Wei, W. Sensitive high-throughput single-cell RNA-seq reveals within-clonal transcript correlations in yeast populations. Nat. Microbiol. 2019, 4, 683–692.
- Saint, M.; Bertaux, F. Single-cell imaging and RNA sequencing reveal patterns of gene expression heterogeneity during fission yeast growth and adaptation. Nat. Microbiol. 2019, 4, 480–491.
- Chetty, A.; Blekhman, R. Multi-omic approaches for host-microbiome data integration. Gut Microbes. 2024, 16, 2297860.
- Ost, K.S.; Round J.L. Commensal fungi in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 723–734.






